Over the past decade, developmental biology entered a golden era, fueled by novel biomimetic approaches to culture human pluripotent stem cells (hPSCs). In vitro recapitulation of the lumenized rosette conformation was described by the group of Magdalena Zernicka-Goetz in 2014 using mice PSCs cultured in micro-wells coated with Matrigel®. This approach was then successfully applied to human ES and iPS cells in 2016 by the same team.
With the view of generating hPSC rosettes at scale while automating their maintenance, several groups have explored droplet microfluidics, confining hPSCs in agarose, alginate or PEG, while other teams engineered microfluidic circuits to entrap and grow hPSCs in 3D. While successfully generating lumenized rosettes, such microfluidic systems were primarily designed for academic research, and thus feature limitations either in throughput or total loading capacity.
Core features of biomimetic approaches
Despite the apparent diversity of in vitro approaches for the generation of hPSC lumenized rosettes, key parameters are generally preserved:
- Cells are biophysically contained and a critical aggregate size needs to be reached to form a polarized epithelium with a central lumen (~50-300 µm).
- Matrigel® remains the standard extracellular matrix, as it mimics the basal membrane niche and supports PSC polarization and lumenogenesis.
- E8 or mTESR feeder-free media are used, minimizing or eliminating the use of poorly characterized biological components additives such as serum, thus avoiding inconsistencies and variations.
- Hypoxic culture conditions (~ 5% O2) are adopted when possible. Hypoxia was indeed reported to reduce mutation rate and promote polarization in hiPSC cultures.
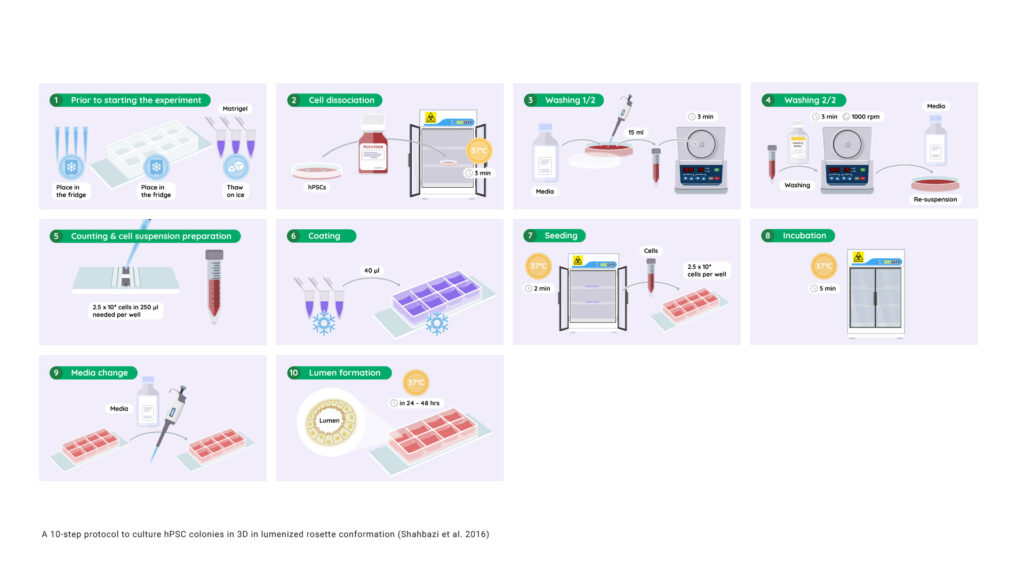
10 steps to obtain lumenized hPSC colonies (Adapted from Shahbazi et al. 2016)
- Place filter tips and ibiTreat 8-well μ-plates in the refrigerator and thaw Matrigel® slowly on ice.
- Detach hPSCs using StemPro Accutase by incubating at 37°C for 3 min.
- Add E8 or mTESR1 and pipette the cell suspension in a 15 ml conical tube. Centrifuge for 3 min at 1000 rpm.
- Wash once with PBS and centrifuge the cell suspension for 3 min at 1000 rpm. Re-suspend the cells in either E8 or mTESR1. NOTE: At this point, ROCK inhibitor may be added to prevent dissociation-induced cell death.
- Count the cells using a hemacytometer. 2.5 ×104 cells are needed per well.
- Coat the entire surface of the pre-chilled ibiTreat 8-well μ-plate with 40 μL of ice-cold Matrigel®.
CRITICAL STEP: Use pre-chilled tips and keep the Matrigel® on ice at all times. Increasing temperatures lead to premature polymerisation. Make sure the Matrigel® covers the entire surface of the well. - Place in a 37 °C incubator for 2 min or until the Matrigel® has solidified. Immediately after, add 2.5 ×104 cells in a final volume of 250 μL per well (cell suspension prepared in 5).
- Incubate the cell suspension for 5 min at 37°C or until most of the cells have attached to the Matrigel®.
- Gently remove the supernatant. Add fresh E8 or mTESR1 medium, supplemented with 5% Matrigel®. NOTE: At this point, ROCK inhibitor may be added.
- Analyze lumen formation 24 or 48 hours after plating.



